P6: Molecular mechanisms of spermatogenic failure at transcriptional and epigenetic level

Nina Neuhaus (neé Kossack), Centre of Reproductive Medicine and Andrology (CeRA) (Homepage)
Sandra Laurentino, Centre of Reproductive Medicine and Andrology (CeRA) (Homepage)
Project summary
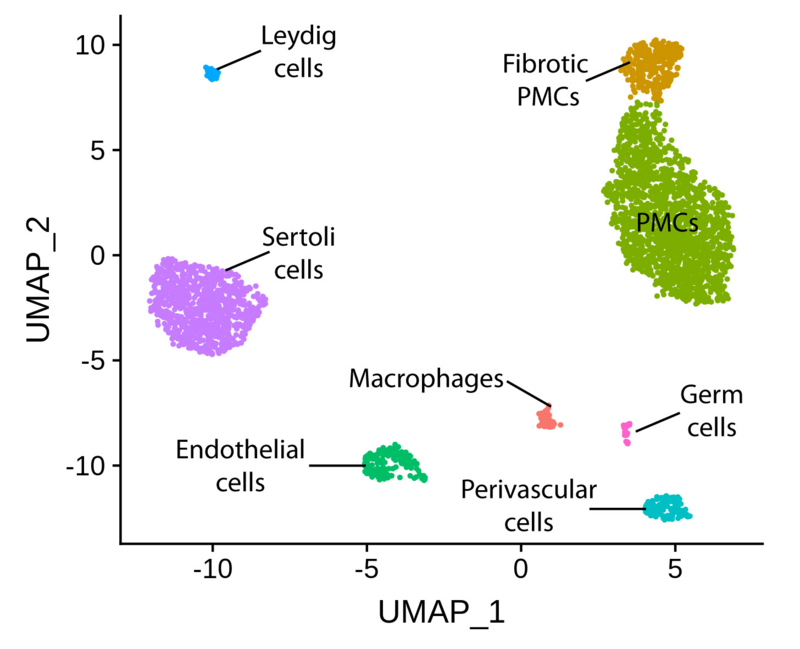
Half of the infertile patients presenting at the CeRA display severely abnormal sperm parameters (oligoasthenoteratozoospermia, OAT). To date, the underlying molecular mechanisms for this phenotype remain largely unknown. Within our CRU project, we have produced high quality methylomes (by whole genome bisulfite sequencing) of germ cells from normal and OAT men. From the same and additional patient groups, we have also generated single-cell transcriptomes (10X Genomics) of testicular cells.
The above mentioned datasets have given us unprecedented insight into the common molecular mechanisms associated with failure of germ cell differentiation in OAT men. Some aberrant expression profiles can be traced back to the germ cell niche and others to spermatogonia, which constitute the most undifferentiated germ cells within the adult human testis.
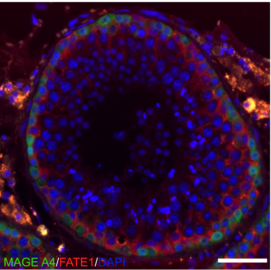
We will use multi-spectral protein imaging to follow up the identified transcriptional changes.
Furthermore, we will increase the number of single cell RNA-Seq datasets to sub-classify the OAT patients according to the altered molecular pathways. Moreover, applying integrated analyses we will assess whether the transcriptional changes are corroborated by alterations of DNA methylation.
Decoding those molecular mechanisms responsible for germ cell failure may enable sub-classification of OAT patients and potentially more precise treatment options in the future.


