Self-Organisation in Cellular Systems
Research Synopsis:Curvature-dependent self-organization in single and collective cell dynamics. To examine formation and function of curvature-induced signaling circuits, we employ cellular (e.g. iPSC-derived human neurons, primary mouse hippocampal neurons, immune cells and HUVECs) and biomimetic model systems that we analyze using advanced microscopy (e.g. lattice light-sheet microscopy, correlative light-electron microscopy, super-resolution microscopy), quantitative image analysis, nanomaterials, biophysical approaches, and numerical modelling. Our ultimate goal is to determine core principles how curvature-dependent self-organization regulates cellular physiology and development in health and disease.
Project 1: Regulation of Cell Dynamics via Membrane Deformations
Goal: Notably, many of the more than 100 curvature-sensing proteins directly change actin polymerization rates, thus altering the force to the PM that initially caused the membrane deformation and triggered their recruitment. The resulting positive and negative feedback-loops are likely employed by cells to control and direct forces that are applied by the cytoskeleton to the PM to organize cellular architecture. How this is coordinated is not known.
Approach: We use advanced image analysis in combination with chemical and gnetic tools to characterize the spatio-temporal localization as well as the functional consequences of curvature-dependent protein recruitment. Currently, we focus on the ~100 proteins with conserved BAR domains.
Significance: Insights from these studies will allow for an exploration of the importance of forces and membrane curvature in a wide variety of cellular processes, including cell motility, collective cell migration, bacterial pathogenesis, viral infection, as well as vesicle endocytosis and fusion.
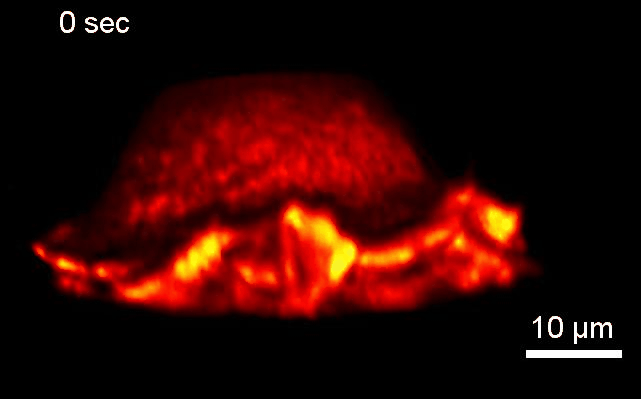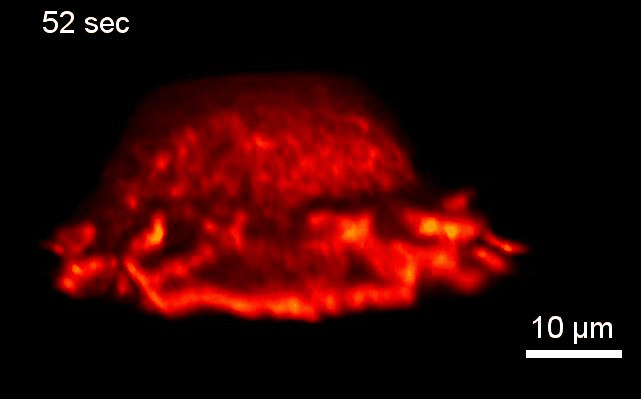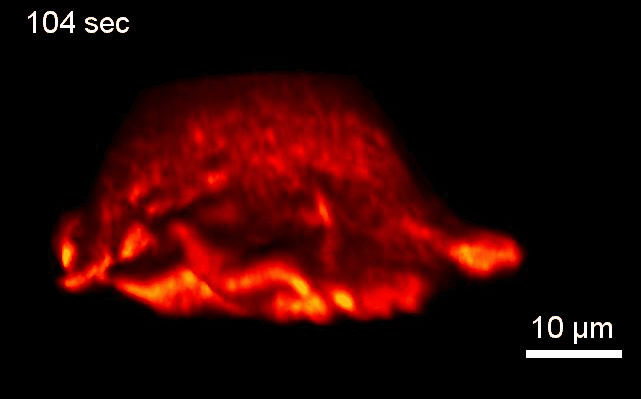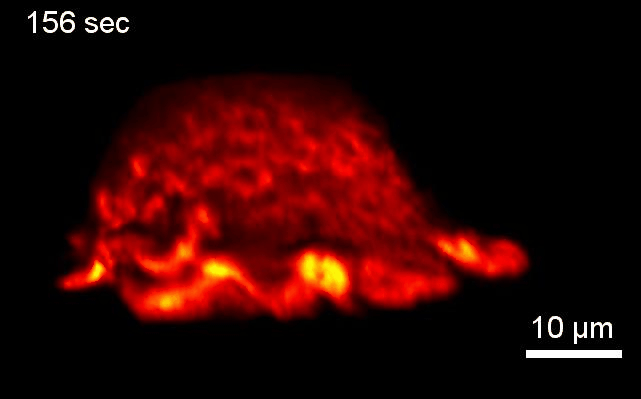
Project 2: Control of Cell Architecture via Membrane Curvature
Goal: We will systematically investigate how neurons sense and coordinate the forces that shape their complex 3-dimensional branched membrane structures. Despite their importance for input integration and signal transmission, the molecular mechanisms that control neuronal architecture remain unclear. Most morphological rearrangements are initiated by changes in actin dynamics near the plasma membrane (PM), and trigger the formation of highly curved membranes. We aim to investigate how the >100 curvature-sensing proteins that directly alter actin polymerization, thus changing the forces applied to the PM affect neuronal architecture.
Approach: Using a genetic and chemical tools we analyse how curvature-dependent recruitment of individual proteins shapes neuronal architecture, as well as how disease-induced changes in PM properties affect curvature-dependent protein recruitment and neuronal architecture.
Significance: This dual ability of selected proteins to sense and modify actin-induced PM curvature is important, and defects of these feedback loops result in neuro-degenerative diseases (eg. OPHN1 and SRGAP2).
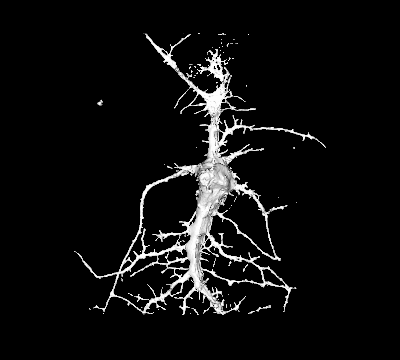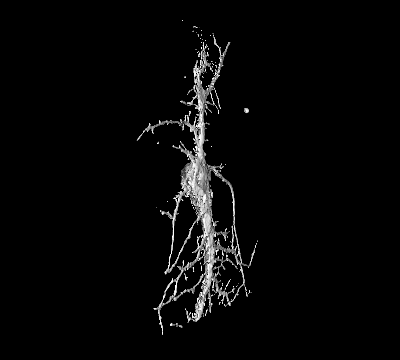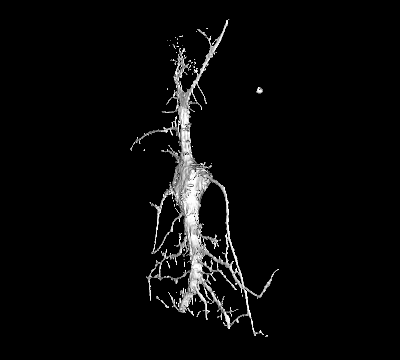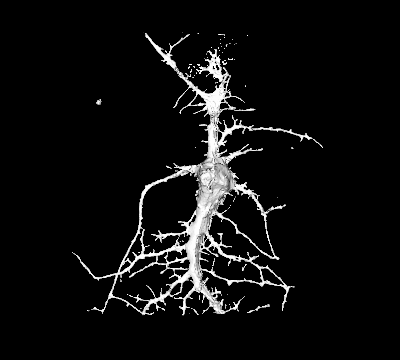
Project 3: Nanostructures to Study Membrane Deformations in Live Cells
Goal: While it is widely accepted that many regulatory mechanisms in cells rely on curvature-dependent membrane interactions, the underlying mechanisms have remained elusive due to the difficulty to studying such processes in living cells.
Approach: We fabricate nanostructures that induce PM curvature to systematically characterize for the first time the biophysical properties of individual curvature-sensing proteins in live cells.
Significance: A segregation of curvature-sensing proteins to subcategories based on spatial (positive or negative curvature, optimal diameter) and temporal (binding affinity, exchange rates, polymer stability) parameters will allow us to characterize the dynamic protein kinetics at the plasma membrane.

Project 4: Tool development
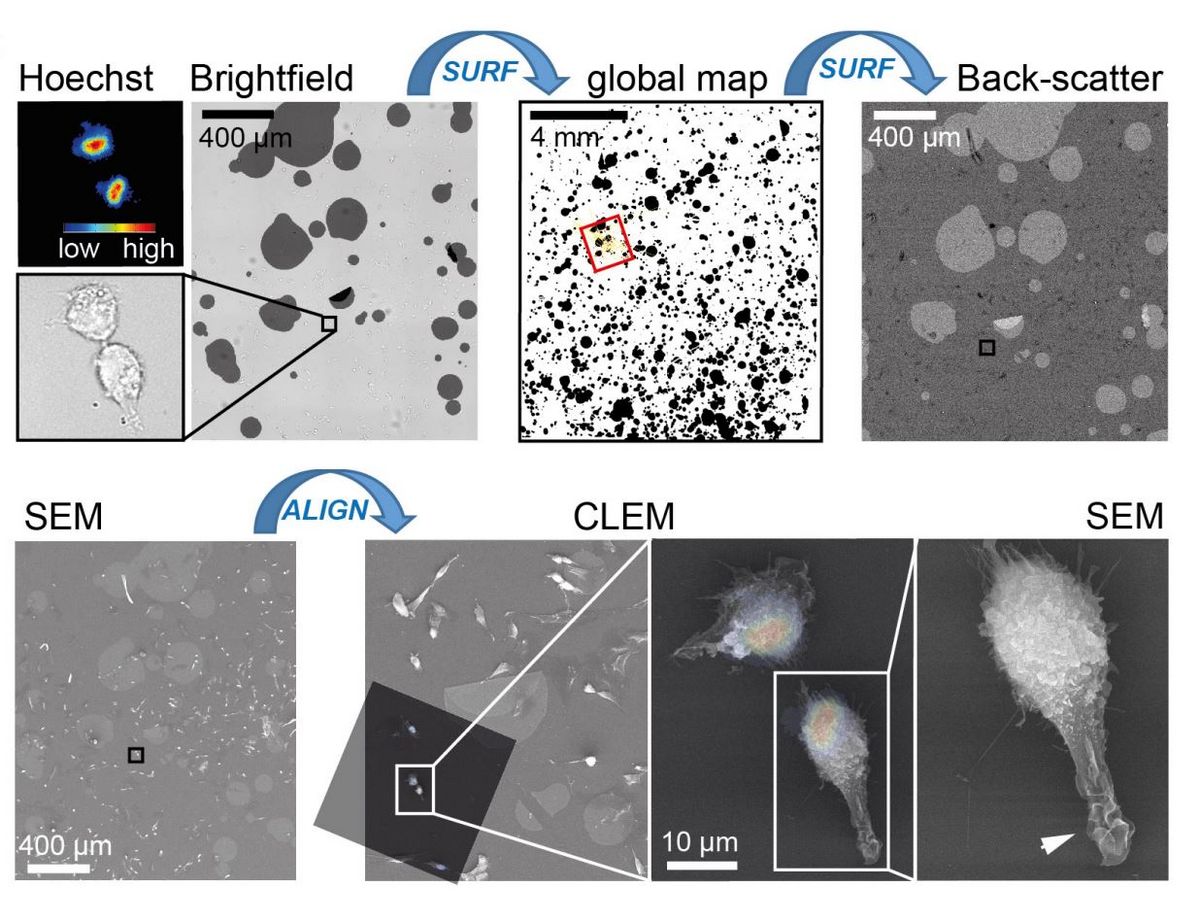
Goal: We develop new approaches to investigate mechanobiological properties of cells. These include among others new approaches for correlative fluorescence-scanning electron microscopy (Fig. 4).
Approach: Our approach combines material science and image analysis tools.
Significance: The development of new approaches facilitates new insights into ultrastructural changes that govern cellular signalling at the plasma membrane.