CP: Bioinformatic pipelines and integrated analyses

Frank Tüttelmann, Institute of Reproductive Genetics (Homepage)
Julian Varghese, Institute of Medical Informatics (Homepage)
(formerly: Martin Dugas, now Institute of Medical Informatics, University of Heidelberg)
Project summary
Identification of genetic variation associated with male infertility relies on the availability of clinical and genetic data from large patient cohorts. Recent technological advances in sequencing techniques not only increased demand for computational resources, but also for in-depth bioinformatic expertise to adequately address each individual OMICs' challenges. While there are established workflows to analyse OMICs data such as bulk RNA-Seq, there is a strong need to optimise bioinformatic pipelines, especially for novel scRNA-Seq techniques. Performing such analyses not only requires application of the algorithms but also validation and benchmarking of results to ensure robustness of conclusions drawn from such data.
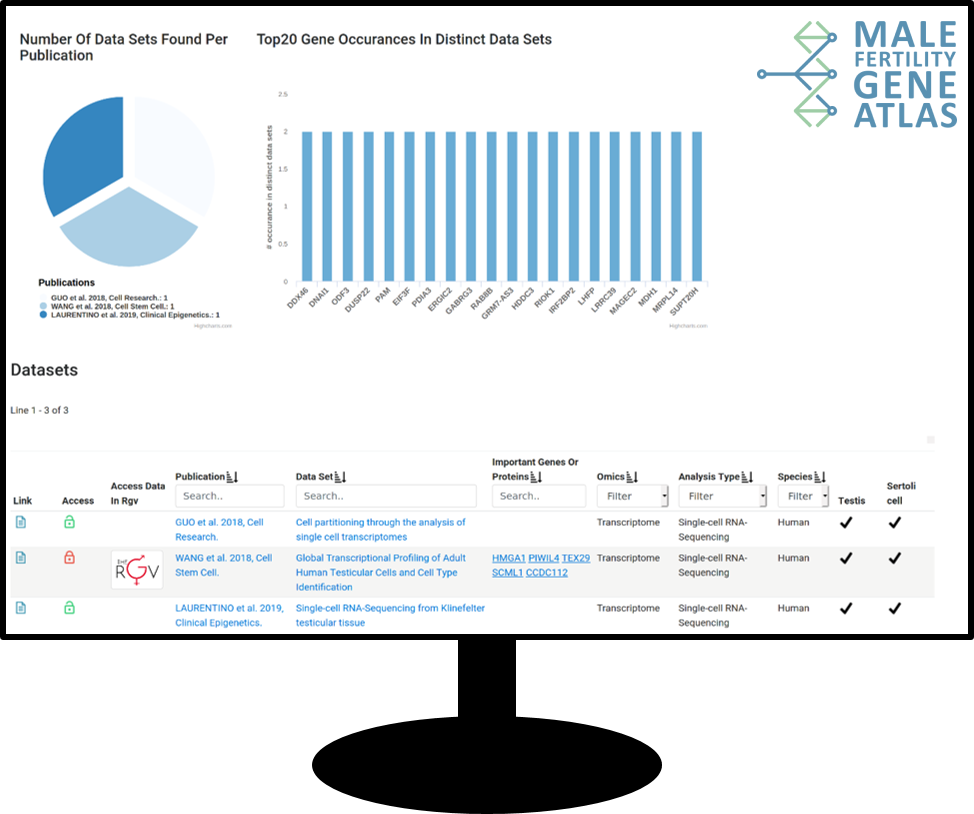
With a centralised Core Project, we provide a high standard of data processing and analysis. In the first funding period, more than 2,900 samples from multiple OMICs (such as whole exome sequencing and (sc)RNA sequencing) have been successfully analysed, ultimately fuelling into the Male Fertility Gene Atlas (MFGA), a comprehensive resource for researchers in the field of reproductive medicine. Consolidating all data in one project further enables application of machine learning techniques to uncover previously unknown associations between biological markers and clinically relevant phenotypes.