Unravelling microbial community features for biomedical applications using graph neural networks
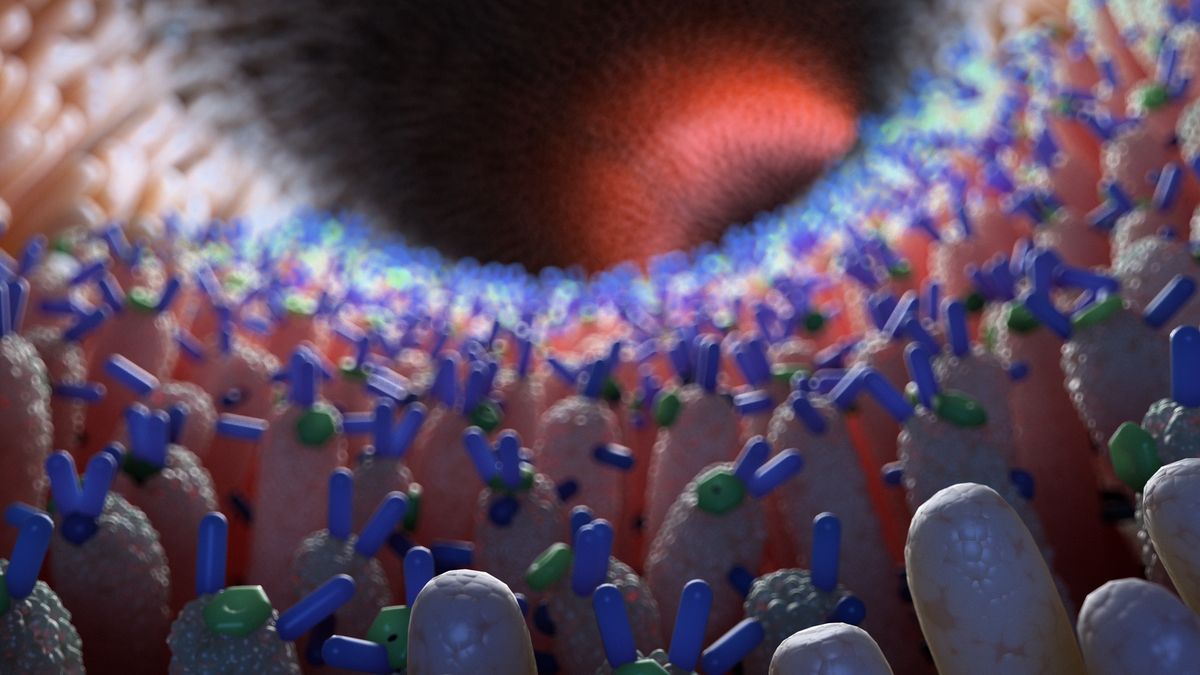
The human microbiome, composed of myriad microorganisms, is crucial for physiology and health; understanding its organization enables new diagnoses and treatments. High-throughput next-generation sequencing (HT-NGS) now characterizes microbial abundance in detail, but microbiome composition varies with genetics, lifestyle, and environment. Microbes also compete and cooperate, forming dynamic communities whose emergent properties influence the host. Assessing all community combinations is technically unfeasible, limiting their use as biomarkers. This project addresses these gaps by integrating Graph Neural Networks (GNNs) into an adapted HT-NGS pipeline to characterize and predict health outcomes from the gut microbiome. Unlike methods for time-series or images, GNNs use graphs, capturing interactions (edges) between microbes (nodes) and integrating diverse data modalities into an interpretable structure.
The project has two complementary aims:
- Investigate gut microbiome diversity and links to metabolic functions across populations, considering lifestyle and genetics, using public datasets from Western societies and Native Amazonian groups;
- Study the microbiome’s relationship with Parkinson’s disease (PD). As <10% of PD cases are purely genetic, environmental and lifestyle factors are key. Previous machine learning models, focused on individual bacteria, yield contradictory results; thus, this project targets bacterial communities as potential PD biomarkers with predictive power.
Contact: Renan Moioli

Funded by the CAPES–Humboldt Research Fellowship


