Stress-induced programming in serotonin (5-HT) system-modified mice: epigenetic editing as a strategy to cope with anxiety
van den Hove, Lesch, Schmitt-Böhrer
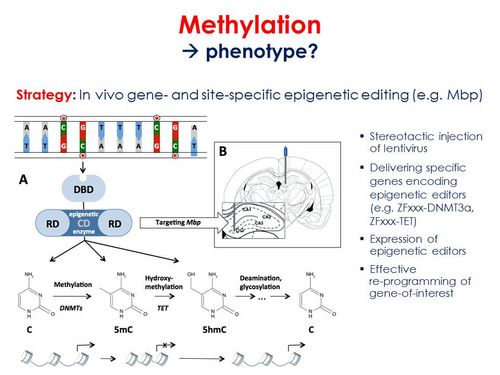
In order to achieve this goal, we will pursue the following objectives:
- Validation of epigenetically regulated risk and protective genes identified in previously established G x E paradigms. We will validate candidate genes derived from the 5-Htt x prenatal stress (PS) paradigm targeting genes that foster resilience to anxiety disorders by techniques which quantify gene expression and differentiate CpG site-specific DNA methylation in a sex-dependent manner and focusing on various brain regions from which specific neuronal cells have been isolated.
- Investigation of epigenetically regulated risk and protective genes validated in Objective 1 in a novel Tph2-deficient mouse model of panic disorder. For this purpose, we will validate candidate genes derived from the 5-Htt x PS paradigm in the Tph2 x PS panic disorder model to elucidate the degree of developmental epigenetic programming of anxiety-related traits. The results are expected to identify pathways underlying inadequate adaptability versus protection.
- Causality assessment and intervention using an in vivo gene- and site-specific epigenetic editing strategy. Zinc finger (ZF) proteins and guide RNAs (CRISPR-Cas9 system) will be designed to target transcriptional activators or repressors to candidate mRNA loci, such as myelin basic protein (Mbp) identified in our 5-Htt x PS study. Making use of stereotactical injections of lentiviruses, specific genes encoding the epigenetic editors (e.g. DNMT3a, TET) will be delivered into the brain (e.g. hippocampus for Mbp) of adult stressed 5-Htt- or Tph2-deficient mice, in order to induce normalize the degree of methylation and expression of the gene of interest (e.g. Mbp) in local neurons. Fear- and anxiety-related behavioural consequences of this approach will be examined, while brain tissue will be assessed for e.g. morphological, neurogenic, epigenetic and transcriptomic changes.
