Special method: Metagenomic sequencing
Our group applies unbiased metagenomic next-generation sequencing (mNGS) to routine and archival neuropathology specimens to improve pathogen detection in infectious and inflammatory diseases of the central nervous system. By combining optimized laboratory workflows for low-input material from formalin-fixed paraffin-embedded (FFPE) tissues and cerebrospinal fluid (CSF) with tailored bioinformatic pipelines, we enable the identification and characterization of bacterial, viral, fungal, and parasitic DNA and RNA directly from diagnostic samples. These approaches complement conventional histology and targeted PCR, facilitate the discovery of unexpected or rare pathogens, and support more precise and timely clinical diagnoses. Our work focuses on the development, validation, and clinical integration of metagenomic sequencing strategies using both short-read (Illumina) and long-read (Nanopore) technologies.
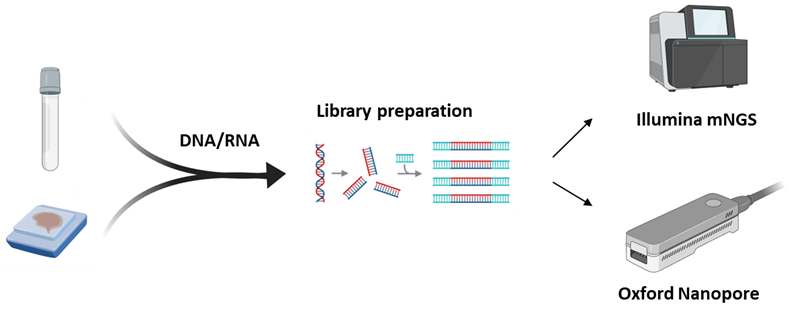
Methodologically, we integrate complementary targeted and enrichment-based strategies to increase analytical sensitivity and spatial resolution. This includes bacterial 16S rRNA gene sequencing from FFPE tissues following guided macrodissection, enabling region-specific microbial profiling within histologically defined lesion areas and adjacent reference tissue and directly linking pathogen detection to morphological context.
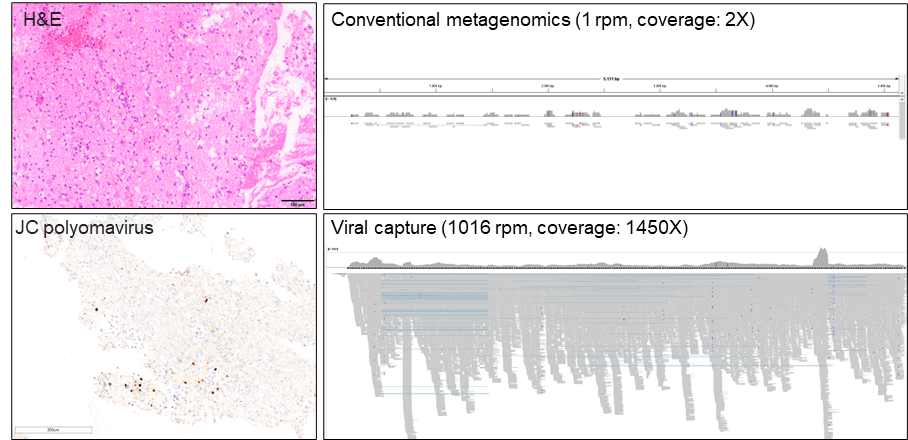
In addition, we employ hybrid-capture–based metagenomic panels that allow the simultaneous enrichment and detection of more than 15,000 viral genomes in parallel, providing comprehensive, high-sensitivity viral screening from low-biomass routine specimens and broad, hypothesis-free pathogen identification in neuropathological practice.
Selected publications
Mohr J, Albers A, Schaumburg F, Paulus W, Brokinkel B, Stummer W, Spille DC, Thomas C. Deep 16S rDNA Sequencing of Chronic Subdural Hematomas Suggests Involvement of Bacterial Infection in Recurrences. Neurosurgery. 2025 Sep 29. doi: 10.1227/neu.0000000000003754. Epub ahead of print. PMID: 41020604.
Dambietz CA, Kintzinger T, Schuler F, Albers A, Suntrup-Krueger S, Fingerle V, Meyer Zu Hörste G, Thomas C. Nanopore sequencing identifies Borrelia miyamotoi as an unexpected cause of meningitis after B cell depletion. Neuropathol Appl Neurobiol. 2024 Dec;50(6):e13017. doi: 10.1111/nan.13017. PMID: 39511958; PMCID: PMC11618485.
Gorißen C, Albers A, Ruf V, Chteinberg E, Siebert R, Schweizer L, Kaufmann L, Kühn JE, Tappe D, Kuhlmann T, Thomas C. Targeted whole-viral genome sequencing from formalin-fixed paraffin-embedded neuropathology specimens. Acta Neuropathol. 2024 Oct 9;148(1):51. doi: 10.1007/s00401-024-02812-z. PMID: 39382575; PMCID: PMC11464609.
Albers A, Spille DC, Suero-Molina E, Schaumburg F, Stummer W, Paulus W, Thomas C. Rapid bacterial identification from formalin-fixed paraffin-embedded neuropathology specimens using 16S rDNA nanopore sequencing. Neuropathol Appl Neurobiol. 2023 Feb;49(1):e12871. doi: 10.1111/nan.12871. PMID: 36534112.
Contact:

Team




Alumni


