Peroxisomal Biogenesis
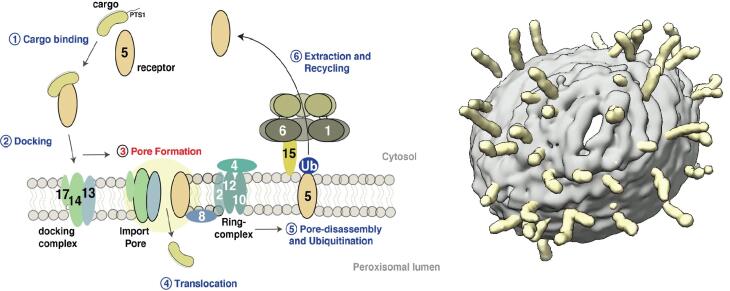
Import of peroxisomal matrix proteins. A general model of the PTS1 import pathway is shown (left image). CryoEM tomography of the peroxisomal docking complex reconstituted in liposomes (right image; from Lill et al., 2020, PNAS) Our research focuses on the molecular understanding of peroxisomal biogenesis. Peroxisomes are dynamic small organelles found in almost all eukaryotes. They are capable of performing a plethora of crucial metabolic functions, including the β-oxidation of fatty acids and the degradation of toxic hydrogen peroxide. The corresponding enzymes are synthesized on free cytoplasmic ribosomes and later imported into the peroxisomal lumen by the peroxisomal import machinery. Peroxisomal dysfunction and impaired peroxisomal import lead to devastating inborn errors of metabolism, further emphasizing the importance of understanding these fundamental biological issues. Along the way, peroxisomes continue to come into focus as signaling platforms in current research and play an essential role in the innate immune response, among other roles. Furthermore, peroxisomes are implicated in a variety of age-associated diseases, including cancer, neurodegenerative processes, and diabetes.
However, the main characteristics of the peroxisomal import process remain poorly understood. Our research aims to characterize and visualize these processes using cryo-electron microscopy (single particle analysis and tomography) and biochemical and biophysical methods to understand this central mechanism in detail.
AAA-ATPases: Biological Nanomachines
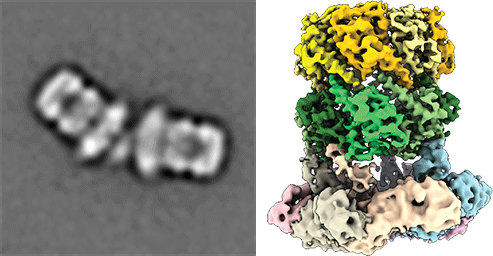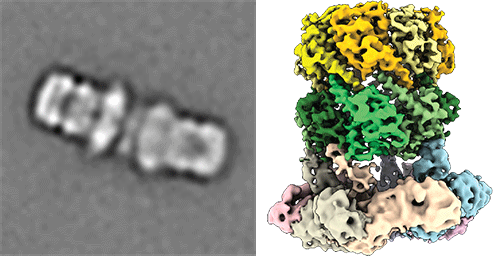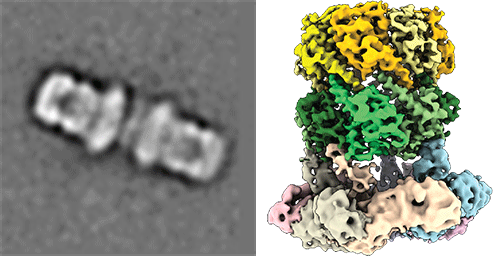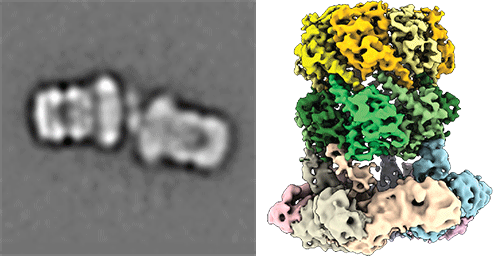
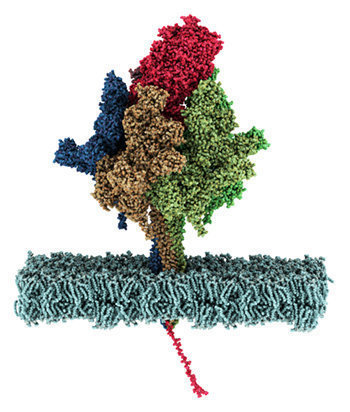
Cryo electronmicroscopic image of the ClpP and ClpX complex from pathegonic bacteria for degradation of proteins. (from Gatsogiannis, Balogh et al., 2019; Nature Strut Mol Biol) Protein quality control systems are important for regulating the balance of the cellular proteome. In this context, the activity and folding state of proteins in all cells is precisely controlled and regulated, especially in the context of stress responses, cell division and virulence of pathogenic microorganisms, among others. AAA+ protease complexes play a central role in this process. For instance, ring-shaped AAA ATPases use the energy from ATP hydrolysis to unfold substrate proteins in an energy-dependent manner. The unfolded proteins can, for example, be passed to proteases for further degradation or recycled to other compartments.
We would like to preserve the structural conformational dynamics of these fascinating molecular machines in action and also directly visualize their interaction network in their native environment. Only then can their functioning be elucidated in detail and thus provide a solid basis for combating serious disorders and diseases that can be attributed to molecular defects in these machines.
Pore-Forming Neurotoxines
The bacterial pore-froming Tc-Toxin (from Gatsogiannis, Lang et al., Nature 2013; Meusch, Gatsogiannis et al., Nature 2014; Gatsogiannis et al. Nature Struct Mol Biol 2016; Gatsogiannis et al. Nature, 2018) Pore-forming proteins represent a unique class of highly specific lipid-binding proteins, which undergo a sophisticated metamorphosis from a soluble prepore, to a transmembrane pore state. We are especially interested to understand the underlying mechanisms of those pore-forming proteins that are capable to insert into membranes, in a regulated manner.
Method Establishment

The development of novel reagents, including nanodiscs and amphipols, has greatly facilitated functional and structural studies of integral membrane proteins (IMPs). We are interested in developing novel and delicate methodologies with the help of the DNA-origami technology, to optimize the sample quality of IMPs for high-resolution structural analysis by cryoEM.
Funded by:

Previous funding:
- 2016-2020: DFG-funded Research Group "PerTrans"
- 2018: JSPS, Uehara Foundation

