Reconstruction of clonal evolution
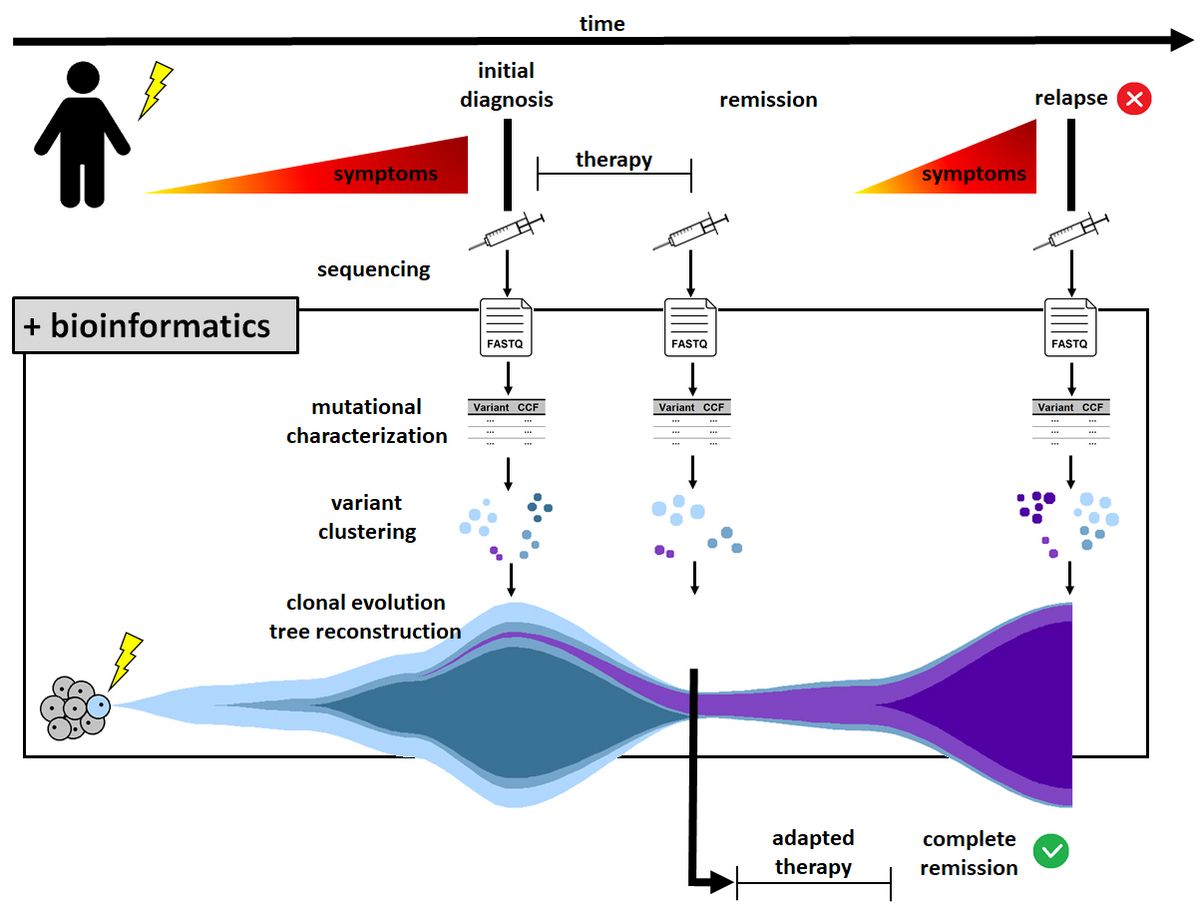
In the field of cancer research, analyzing the clonal evolution of a tumor is of major interest. The early identification of aggressive subclones can enable new therapeutic options. Therapy resistance might be detected prior to relapse, thus, having the potential to influence survival significantly. However, the actual reconstruction of clonal evolution on the basis of mutational data is not trivial, but a challenge for bioinformatics.
We currently have collaborations in the following clinical and theoretical research projects:
Analysis of clonal evolution in adult and pediatric cases of T-lymphoblastic lymphoma, adult and pediatric cases of T-cell acute lymphoblastic leukemia; cooperation with the Department of Paediatric Hematology & Oncology of the UKM (Univ.-Prof. Birgit Burkhardt) and the Department of Hematology of the University Hospital Schleswig-Holstein, Kiel (Prof. Monika Brüggemann)
Analysis of clonal evolution in pediatric cases of myelodysplastic syndrome and acute myeloid leukemia; cooperation with the Medizinische Hochschule Hannover (Dr. Yvonne Behrens)
Optimization of algorithms for automatic reconstruction of clonal evolution; cooperation with the Institute of Computer Science of the WWU (Univ.-Prof. Xiaoyi Jiang)
Analysis of clonal expansion in the human male germline; cooperation with the Clinical Research Unit „Male Germ Cells: From Genes to Function“ (Univ.-Prof. Schlatt and PD Dr. Nina Neuhaus)
Results of our work, analyzing clonal evolution, have been published:
Visualization of clonal evolution + automatic estimation of additional time points and therapy effect with the tool „clevRvis“ (https://academic.oup.com/gigascience/article/doi/10.1093/gigascience/giad020/7113330, https://www.bioconductor.org/packages/clevRvis)
Systematic evaluation of tools for the automatic analysis of clonal evolution, based on simulated data + simulation tool „clevRsim“ (https://www.mdpi.com/1660-4601/20/6/5128/html, https://github.com/sandmanns/clevRsim)
Clonal evolution in multiple myeloma (https://www.frontiersin.org/articles/10.3389/fonc.2022.919278/full)
Clonal evolution in pediatric myelodysplastic syndroma and acute myeloid leukemia (https://www.frontiersin.org/articles/10.3389/fonc.2022.888114/full)
Evaluation of tools for the automatic analysis of clonal evolution, based on exemplary real data (https://cgp.iiarjournals.org/content/19/2/194.long)
Clonal evolution in Burkitt lymphoma (https://www.nature.com/articles/s41375-020-0862-5)
Clonal evolution in adult myelodysplastic syndrome (https://www.nature.com/articles/ncomms15099)
Contact: PD Dr. Sarah Sandmann


