Submissions
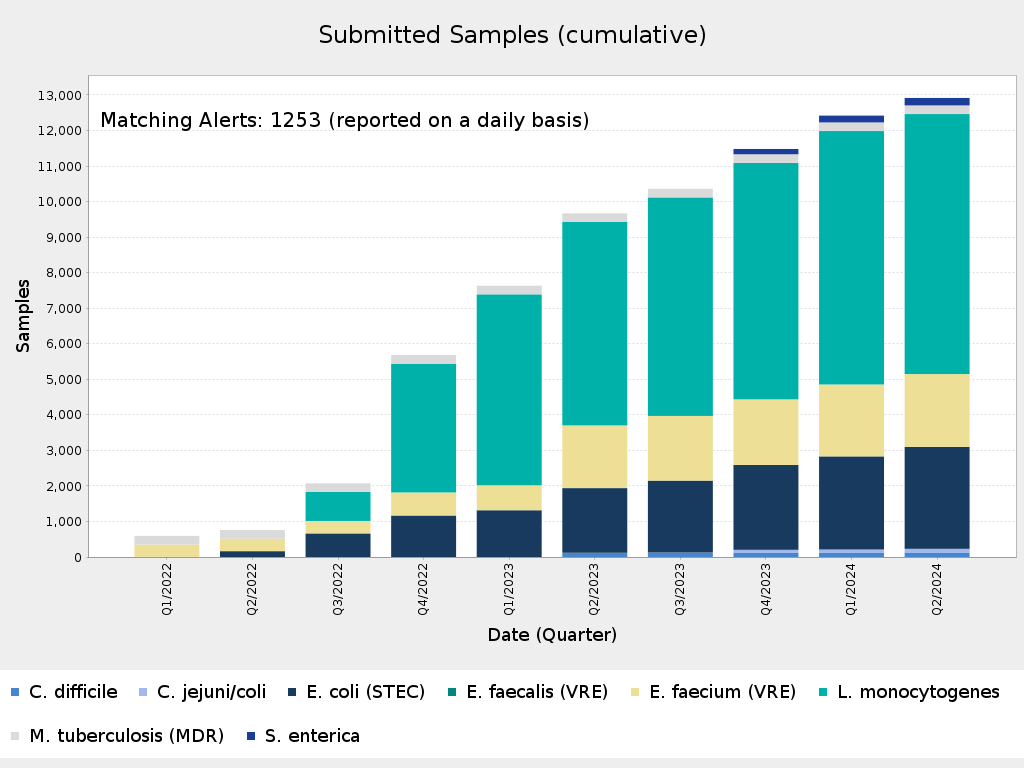
Total Number of Samples/Sequences in the miGenomeSurv Database * powered by Ridom SeqSphere+
Nomenclature
Genomic nomenclature of microbes
A European vision paper that was endorsed following the German 2011 EHEC crisis stated that as a number one priority “a common nomenclature for molecular typing data“ is needed. Prerequisites for such a universal genome-based nomenclature are that it scales with arbitrarily large datasets, is additive and expandable, and that the computing costs for adding new genomes is kept low. Therefore, the miGenomeSurv consortium employs a de-centralized genome-wide gene-by-gene allelic comparison approach that fulfills those prerequisites. By extending the classical multi-locus sequence typing (MLST) approach to the genome level, genomes are compared based on genes common to all isolates, called the ‘core genome’. Thereby a core genome MLST (cgMLST) procedure, based on a fixed and agreed upon number of genes for each species, is well suited for standardizing WGS-based microbial genotyping among various laboratories.
powered by Ridom SeqSphere+
Approach
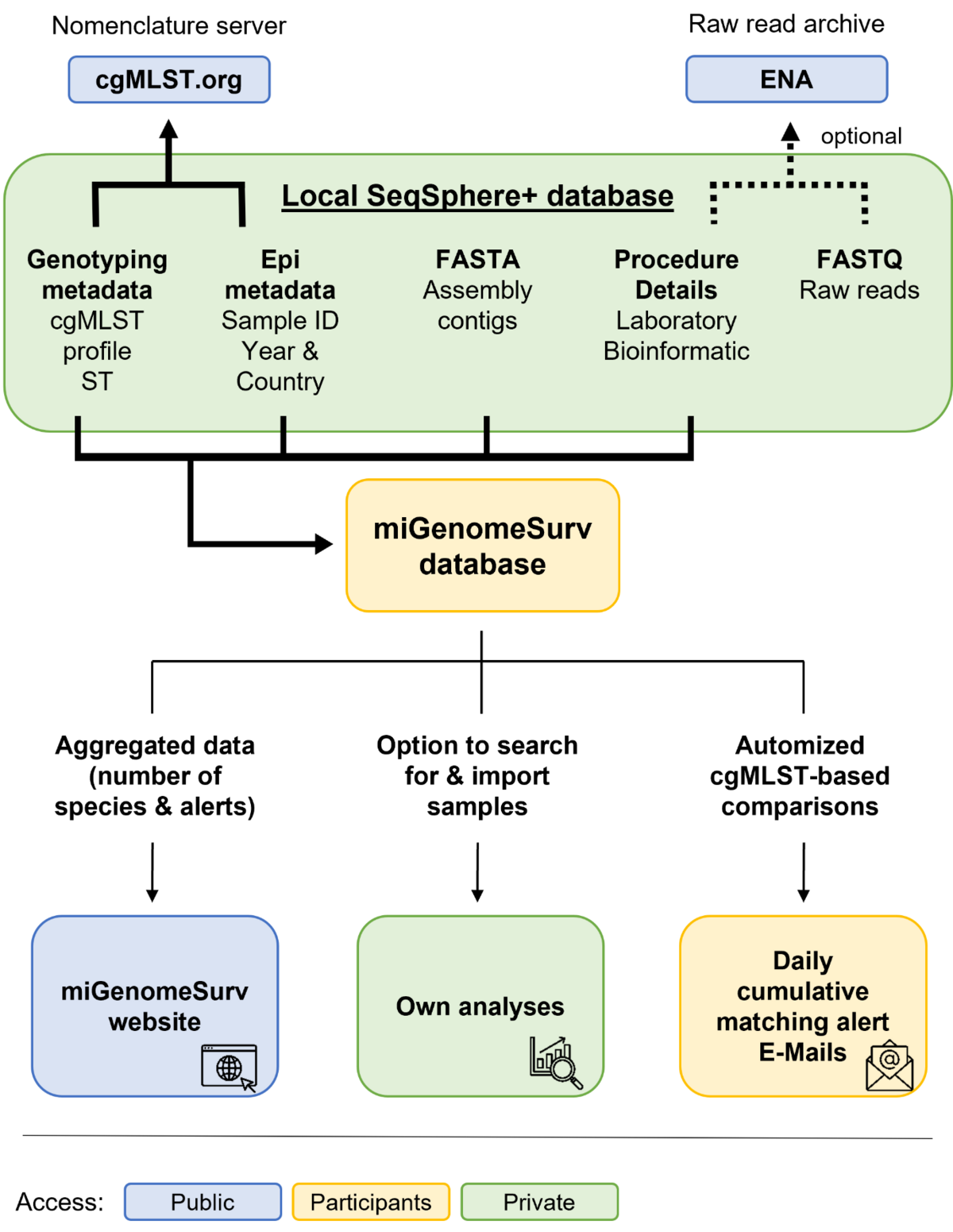
The miGenomeSurv consortium employs a de-centralized cgMLST approach. This means all extensive computing work (e.g., assembly, allele calling, etc.) is done locally on the computers of the consortium partners and the very large raw read data (FASTQ files) are saved locally. The miGenomeSurv data flow comprises elements for public and non-public, i.e. restricted, use (see figure).
To collate the consortium partners data, all partners must submit at least all new allele sequences for allele number assignment to the public and central cgMLST.org nomenclature server (see figure below). All partners may also submit their FASTQ files for raw data preservation (e.g., for publication and/or backup) to public servers like European Nucleotide Archive (ENA). Allelic profiles and the FASTA assembly contigs of the microbial samples together with the submitter contact details, some of the genotyping and epidemiological metadata (only sample ID, year and country of isolation) and some details regarding laboratory and bioinformatics procedures for quality control and assurance are submitted to the non-public miGenomeSurv database. This database with restricted access automatically triggers outbreak early warnings by sending emails to all data submitters in case of close genomic matches between samples. The miGenomeSurv website is meant to inform the public and contains among others aggregated sample data from the miGenomeSurv database.
Are you interested in contributing your genomic data to the consortium? Please contact us for further details.
powered by Ridom SeqSphere+